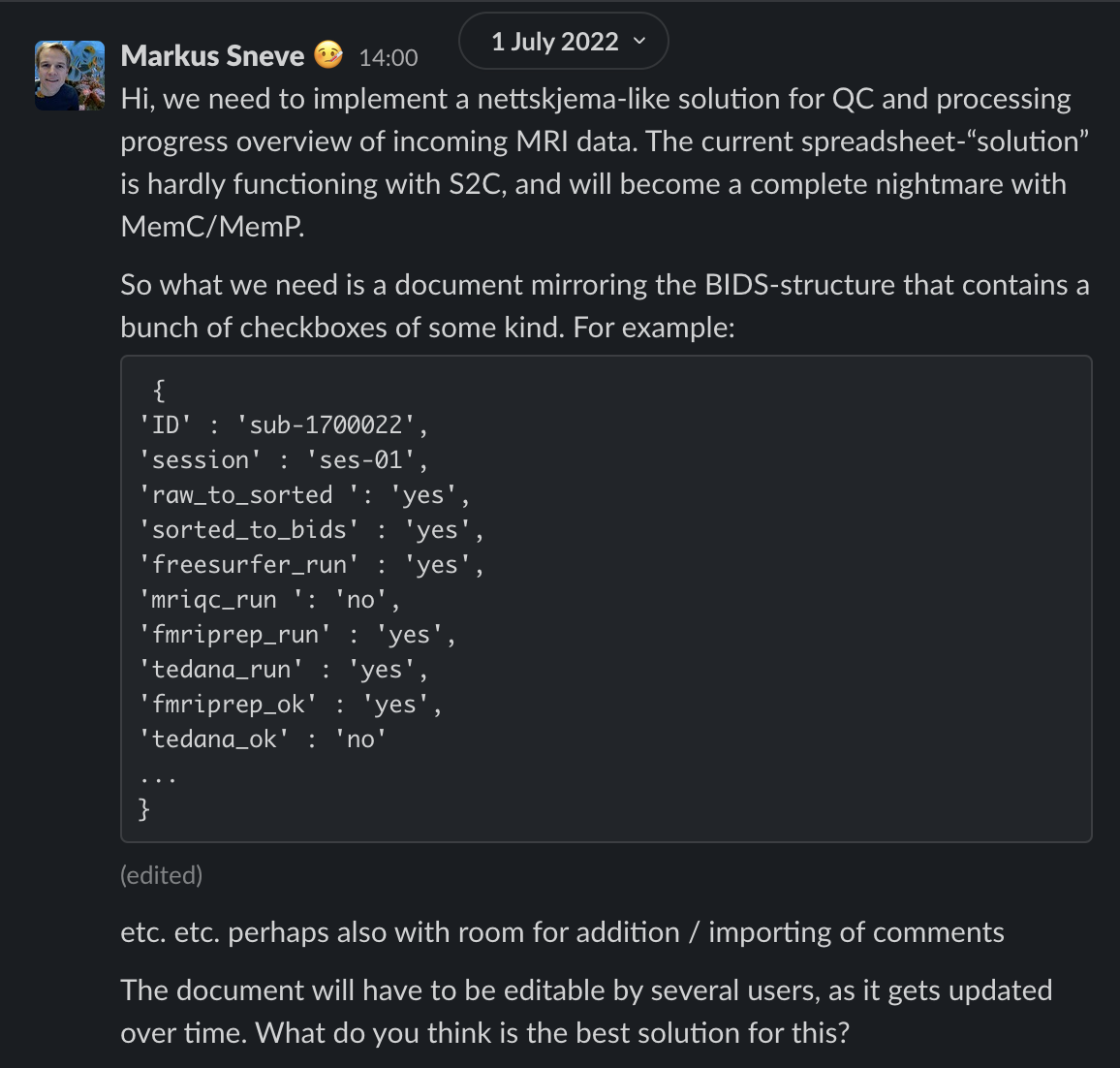
Custom MRI processing status overview
– Keeping track of LCBC’s MRI pipeline –
When a colleague asks for help. ![]()
You jump
What is the issue?
How do we keep track of the MRI data?
- What is the expected processing pipeline for a specific dataset?
How do we know what are possible issues with an MRI dataset?
Where do we look for this information?
- How easily can new and existing staff find the information they are after?
How can we update this information?
How do we keep track?
Problem
MRI has many processing steps
Many steps must be done in a serial fashion.
Knowing which step is next can be difficult
Knowing if any steps have failed and why is difficult
Need
A system that:
- tracks step progression
- indicates expected pipeline to follow
How do we know possible issues with a dataset?
Problem
MRI data are highly annotated
Issues at scanner
Issues in import
Issues in processing
Stored project-wise in spreadsheets
- Can only be edited by one person at the time
Need
A system that:
Gathers all annotations to the same place
Classifies them into meaningful categories
- i.e. belonging to certain modularities
Makes it possible to edit annotations
Where is this information?
Problem
Project-specific logging
distributed in many files
no logic in location
Stored in spreadsheets
Only one can edit
Gets locked and then copied
Each has unique organisation
Need
A system that:
Collects all logs for all projects
Has a standardised way of storing data
- also allowing for project specific setups
Allows multiple users to interact with it
How do we update?
Problem
Updates to the logs are 100% manual
Who does it?
Who has the newest information?
How do they do it?
What is the newest information?
Need
A system that:
allows manual and automatic update of information
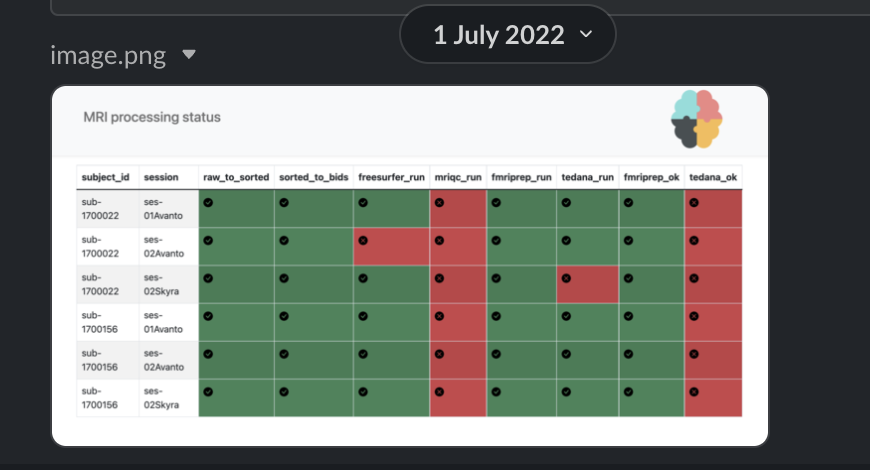
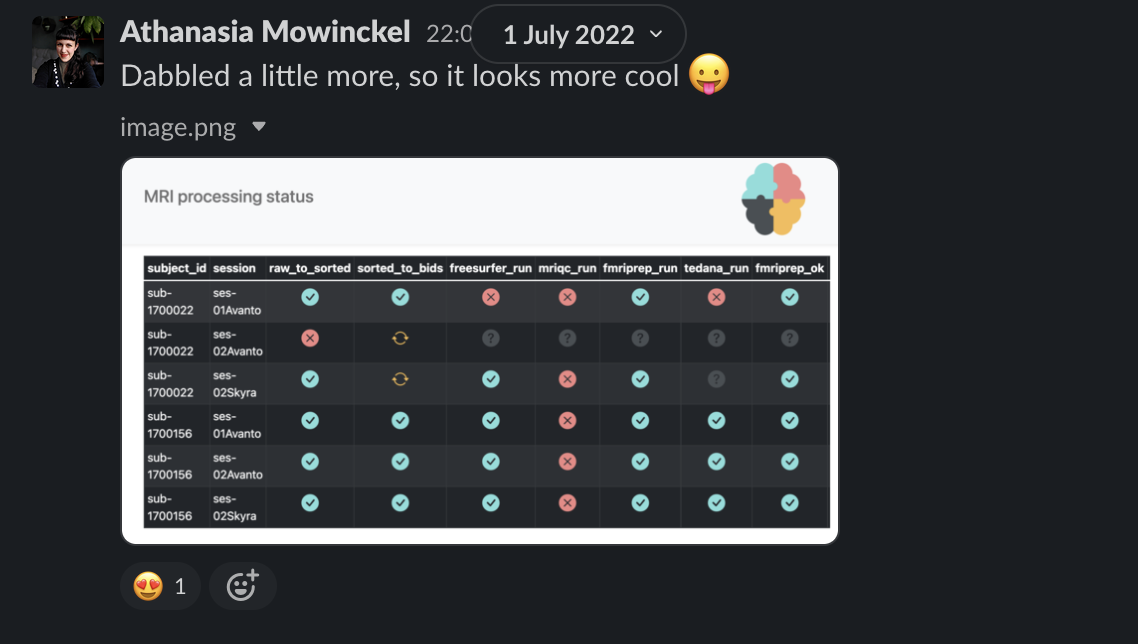
creates easy visual representations to check what has been done
is the single source of needed information
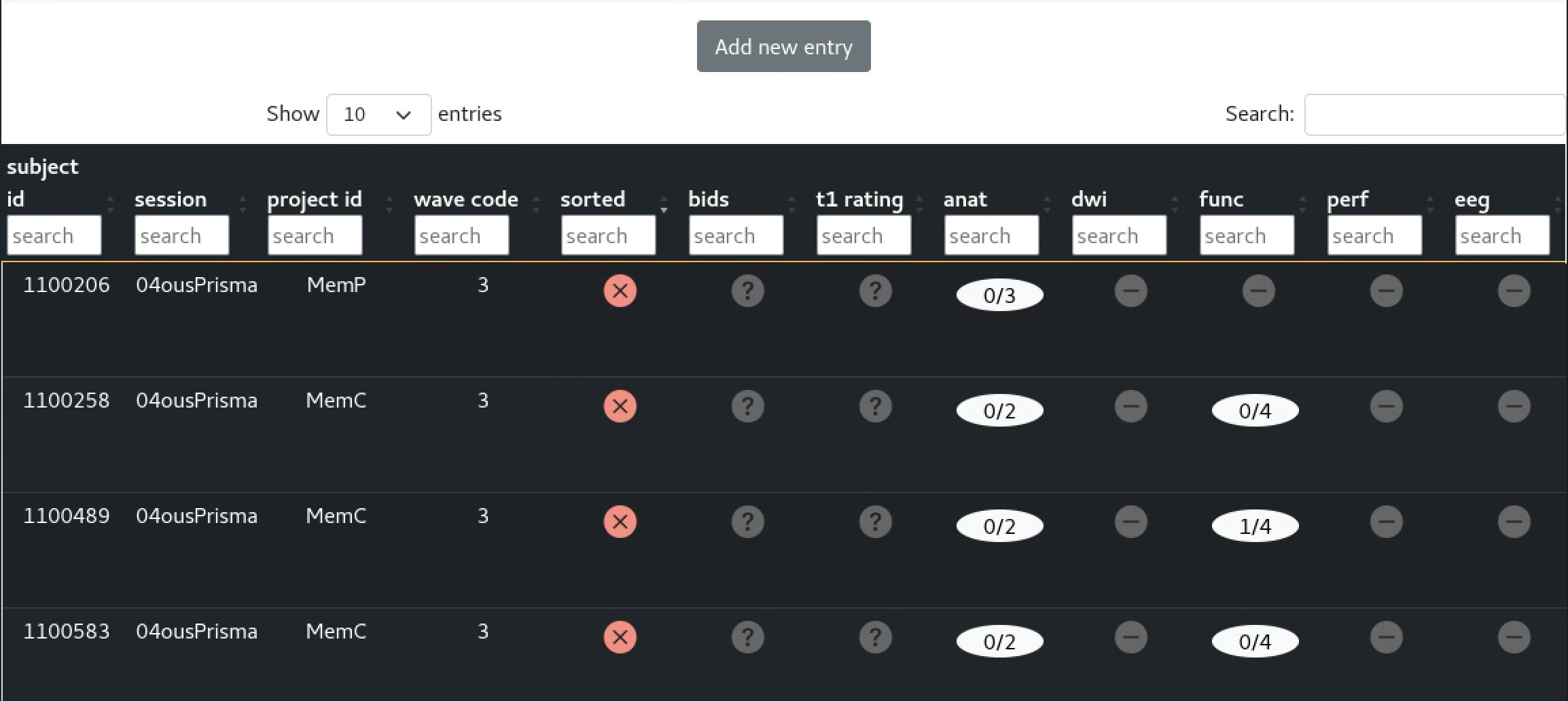
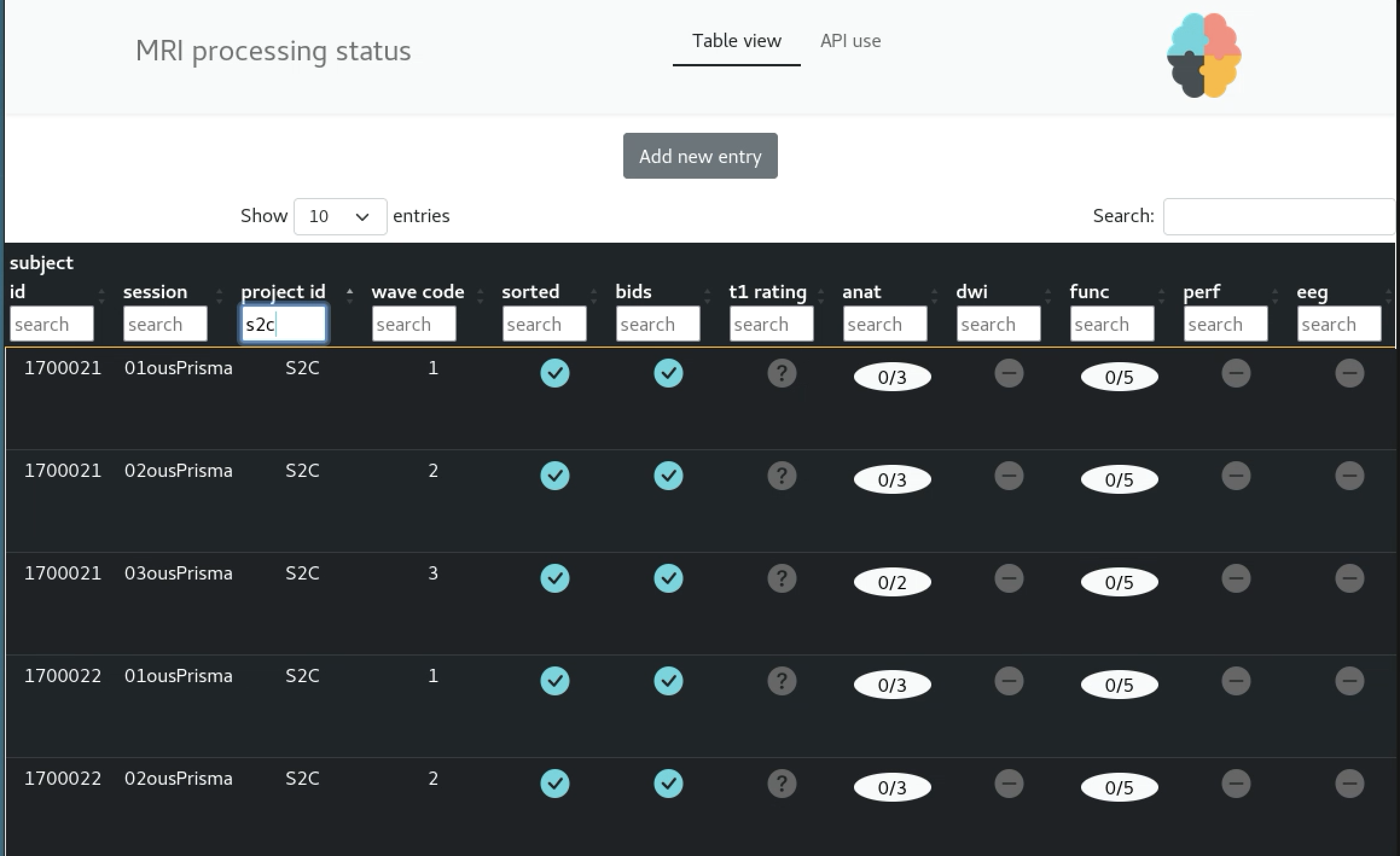
The current solution
Video of the UI
Video of the API
The underlying data - Manual editing
The underlying data - Manual editing
tasks.json
shown on the webui pop-up
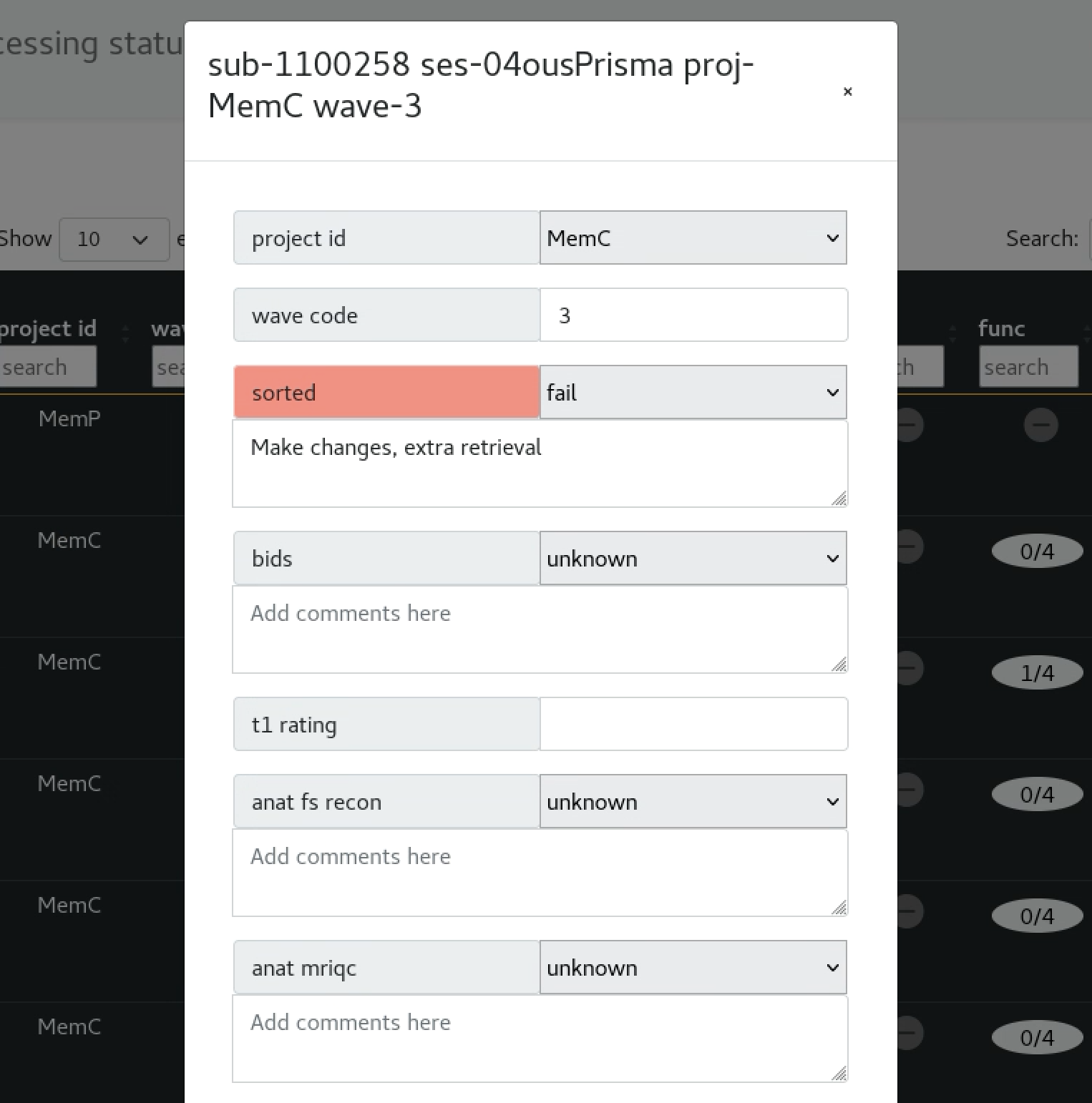
The underlying data - Manual editing
Future work
Application
API endpoints
task.json
protocol.json
Streamline code more
Switch from GET to POST where appropriate
Pipeline integration
Get comments and ratings from nettskjema
Integrate with MRI rating application
Have pipelines update the jsons when tasks finish/fail
Populate for all legacy data
All these and more listed on GitHub